Large-scale docking for drug design on Azure
“Only 10 total drugs in 46 years have been intentionally developed for childhood cancer and have reached FDA approval (reference). Childhood cancer will affect 1,900,000 adults in North America this year, but only 16,000 children. The most common cancers in children often only account for 400-500 of those 16,000 pediatric cases, which is the reason childhood cancers do not pencil out as markets for pharmaceutical companies. The paradox is that childhood cancers are simpler, and often only have a single mutant protein (reference), and computer modeling of chemicals that bind these proteins can lead quickly to new drugs for children with cancer. “
-- Charles Keller - Scientific Director, Children's Cancer Therapy Development Institute
Charles indicates that one of the major challenges in drug design is the scarcity of computing power. Docking is the method to estimate how a molecule attaches to a protein, and it helps to discover possible drug candidates, but it also requires a lot of trial and error and computational resources.
This article describes proof of concept of docking simulation on Azure for “rhabdomyosarcoma”, which is the most common type of soft tissue sarcoma in children. The target is a PAX3-FOXO1 fusion protein that is an essential initiator of rhabdomyosarcoma. Autodock Vina will be used to find compounds poses interacting with the fusion protein with good binding score.
Dataset
Tyuji Hoshino (星野 忠次)’s research team, Laboratory of Molecular Design of Chiba University, have been working with Charles to perform the docking screening to find candidate compounds. The dataset they are targeting, “Namiki_2019”, contains ~4.8 million compounds. Since the number of compounds is large, researchers separate those compounds into batches for processing. The top thousands of compounds in the binding scores by Autodock Vina will then be re-evaluated for the next step of drug design. Usually, the process of docking simulation would take months or even years to complete.
We choose 1,020 compounds out of the total compounds (#3,988,000 ~ #3,989,019) for the proof of concept. This size is suitable enough for initial findings of the nature of the simulation, particularly the CPU, memory, and I/O utilization. As well as finding out the most cost-effective SKU of Virtual Machines to be used, and estimating the total elapsed time when simulating the whole set of compounds.
Architecture
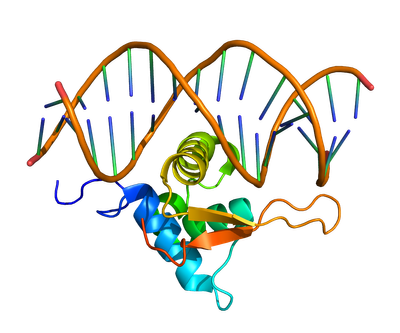
The diagram in Figure 2 illustrates the high-level classic architecture of an Azure High Performance Computing (HPC) environment. The compounds to be simulated are securely stored in a 4TB volume on Azure NetApp Files (ANF) with Standard tier. The simulation job is initiated by a PBS scheduler housed on a CycleCloud server. CycleCloud will dynamically scale up and down, provisioning parallel VMs with the required number of VMs to run the Vina simulation. These VMs are part of a VM scale set (VMSS) with accelerated networking, ensuring optimal performance. Upon completion of each simulation, output files are written back to the ANF volume for secure storage and easy access. This architecture leverages the power of cloud computing to provide a scalable and efficient solution for running complex simulations like Autodock Vina.
Nature of the docking simulations
Most docking tools like Autodock Vina supports multithreading to speed up the simulation by taking advantage of multiple CPUs. We first did some pre-work to find out the most cost-effective # of threads (CPUs) per compound. We also compared two SKUs of Virtual Machines, one is Standard_HB120rs_v3, which feature 120 AMD EPYC™ 7V73X (Milan-X) CPU cores, the other is Standard_D64d_v5, which features the 3rd Generation Intel® Xeon® Platinum 8370C (Ice Lake) processor. Both with Ububntu 20.04. We tested several rounds of different # of CPUs on different compounds to find out the nature of behavior, and below shows one of the results (compound #3,985,126).
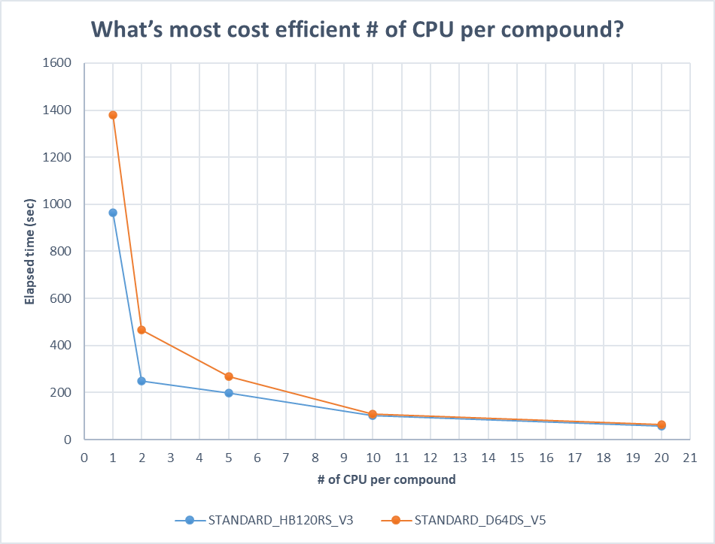
We found that CPU=2 is the most cost-effective setting for all the tested compounds, and HBv3 series is more performant than Dv5 series. We also found that Autodock Vina docking simulation is very compute-intensive with very less I/O utilization. We also found it’s utilizing ~10GB memory per compound, which implies L3 cache size and memory bandwidth could be critical to overall performance. HBv3 also has lower price per CPU compared to Dv5 series. We also found docking simulation is embarrassingly parallel and can be divided into completely independent Vina jobs. Considering all the above we choose CPU=2 and HBv3 as our compute nodes.

Simulations configuration
Below the PBS command we used to submit the Vina jobs:
This command will submit Vina jobs to the PBS queue, each requesting 1 node of type “hb120v3” with 2 CPUs, and allowing the system to freely allocate the resources. Below is the screenshot of CycleCloud portal after submitting the jobs. As we configured to run on 2 CPUs per compound, up to 60 compounds can be run in one HB120v3 VM. Therefore, there were17 HB120v3 VMs provisioned in total, which contains 2,040 cores.
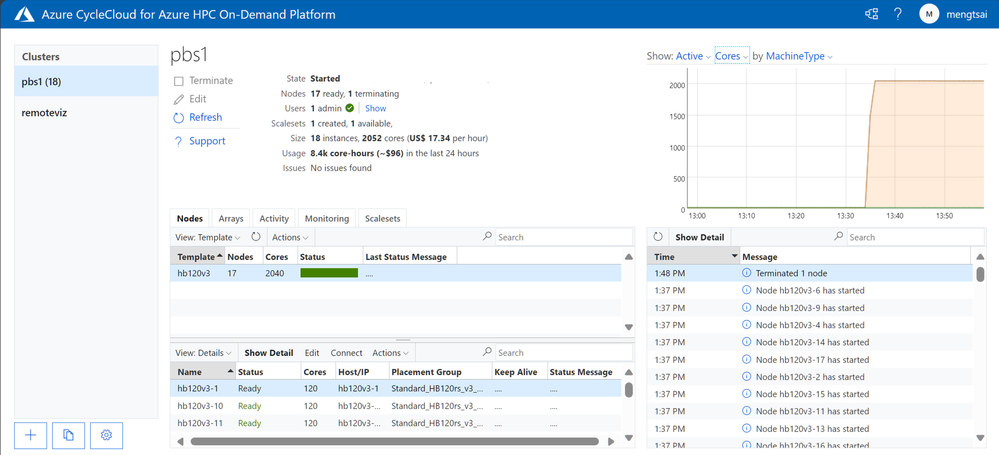
Results and observations
Professor Tyuji helped confirm the predicted docking poses of the simulation are all correct.
Below shows the CPU utilization of one of the HB120v3 VMs. We found CPU usage stays high during the simulation, and around 90% of compounds are done in10 mins. All 1,020 compounds are completed in ~30 mins.
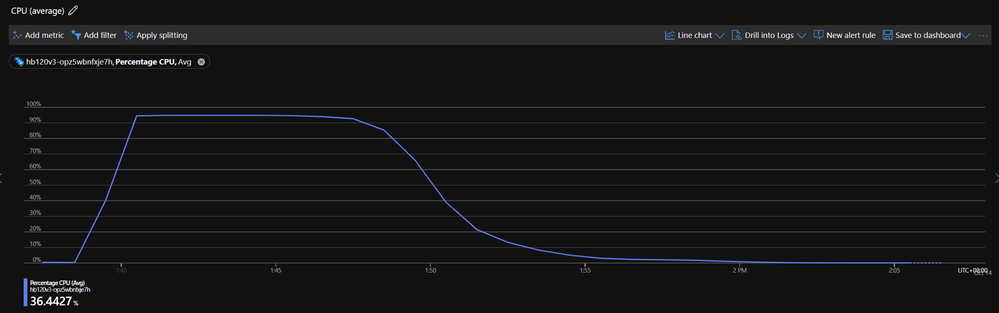
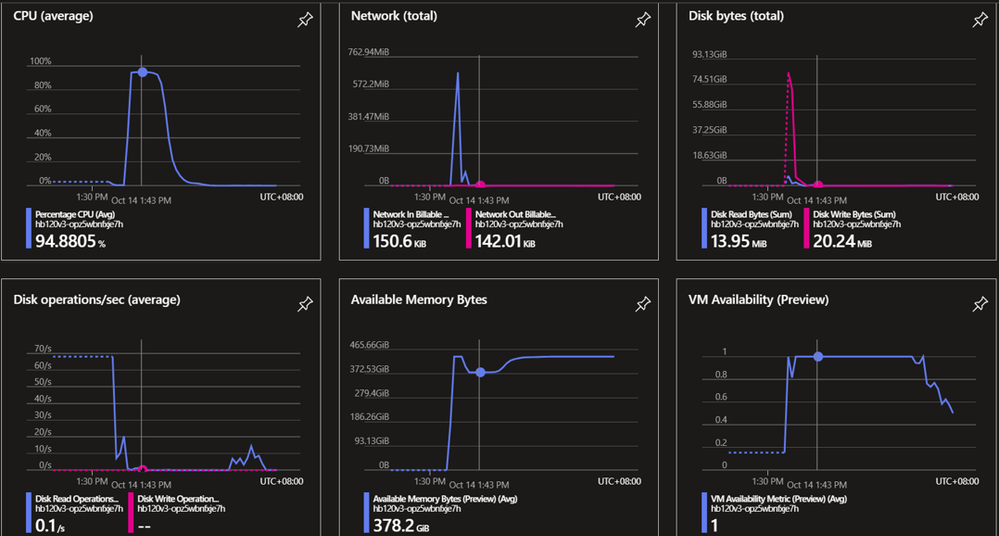
Figure 7. shows more detailed metrics. Besides the high CPU utilization and ~60GB memory usage in peak, we see very little I/O or networking usage. That means customers might not need to use ANF, instead, a proper size of Azure premium disks would be enough to fulfill the requirements with lower cost without affecting the overall performance.
The total size of the output files is ~40MB after screened 1,020 compounds.
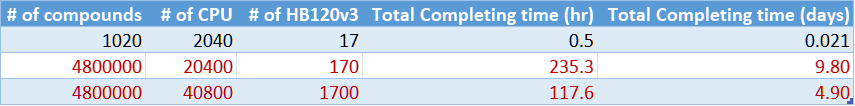
As stated, Azure HPC environment with CycleCloud enables users to easily scale up to 170 or even larger numbers of VMs. Figure 8 shows the estimation (in red) when running ~4.8 million compounds on the larger scale environment. Which shows the total simulation computation time can be completed in 10 days or less, instead of months or years. Please note the estimation is not counting other efforts like infrastructure preparation and data movement time.
Summary
This proof of concept found out the nature of the behavior of running Autodock Vina on the selected dataset, which is very compute-intensive with very less I/O utilization, and ~10GB memory per compound. We found CPU=2 the most cost-effective and HBv3 the suggested SKU. Most importantly we verified the classic Azure HPC environment can run Autodock Vina simulation seamlessly with great scalability. We also suggested a more cost-effective storage solution. And finally, we forecasted an estimation of completion time when running ~4.8 million compounds based on all those findings.
References
High-performance computing (HPC) on Azure: High-performance computing (HPC) on Azure - Azure Architecture Center | Microsoft Learn
What is Azure CycleCloud? Overview - Azure CycleCloud | Microsoft Learn
Published on:
Learn moreRelated posts
Microsoft Purview: Data Lifecycle Management- Azure PST Import
Azure PST Import is a migration method that enables PST files stored in Azure Blob Storage to be imported directly into Exchange Online mailbo...
How Snowflake scales with Azure IaaS
Microsoft Rewards: Retirement of Azure AD Account Linking
Microsoft is retiring the Azure AD Account Linking feature for Microsoft Rewards by March 19, 2026. Users can no longer link work accounts to ...
Azure Function to scrape Yahoo data and store it in SharePoint
A couple of weeks ago, I learned about an AI Agent from this Microsoft DevBlogs, which mainly talks about building an AI Agent on top of Copil...
Maximize Azure Cosmos DB Performance with Azure Advisor Recommendations
In the first post of this series, we introduced how Azure Advisor helps Azure Cosmos DB users uncover opportunities to optimize efficiency and...
February Patches for Azure DevOps Server
We are releasing patches for our self‑hosted product, Azure DevOps Server. We strongly recommend that all customers stay on the latest, most s...
Building AI-Powered Apps with Azure Cosmos DB and the Vercel AI SDK
The Vercel AI SDK is an open-source TypeScript toolkit that provides the core building blocks for integrating AI into any JavaScript applicati...
Time Travel in Azure SQL with Temporal Tables
Applications often need to know what data looked like before. Who changed it, when it changed, and what the previous values were. Rebuilding t...